Skimming Step for RECASTing the VHbb Analysis
Overview
Teaching: 10 min
Exercises: 15 minQuestions
How do I use the yadage syntax I’ve learned to preserve the analysis steps needed to prepare my signal for interpretation?
Objectives
Practice using yadage syntax to describe the first two steps of our VHbb analysis.
Add a new executable to your analysis framework to convert the ROOT histograms created by Analysis payload to a simple text file format.
Introduction
We now have all the yadage tools to put together our VHbb RECAST workflow, starting by “yadage-ifying” our first analysis step using the syntax we just learned. We can use essentially the same yadage structure and syntax for defining our analysis steps as we did for the message writing and shouting steps.
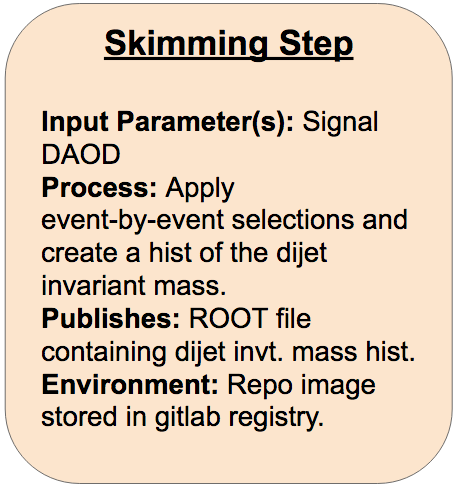
Skimming Step
On gitlab, create a new repo to contain your workflow. Name it something like danika-workflow (but with your name instead of mine). Clone your new repo onto your computer.
In another shell, cd into the workflow repo and start the yadage container so you can validate and test the steps and workflow as you develop. You’ll also need to log in to the gitlab docker registry using your CERN credentials so yadage can automatically pull images from the gitlab registry:
cd [your workflow repo]
docker run --rm -it -e PACKTIVITY_WITHIN_DOCKER=true -v $PWD:$PWD -w $PWD -v /var/run/docker.sock:/var/run/docker.sock yadage/yadage sh
docker login gitlab-registry.cern.ch
Back in your first shell, cd into your new workflow repo, and create your empty steps.yml and workflow.yml files. Next, create a directory called inputdata, copy your signal DAOD file into inputdata, and rename it recast_daod.root:
cd [your workflow repo]
touch steps.yml
touch workflow.yml
mkdir inputdata
cp /path/to/DAOD_EXOT27.17882736._000008.pool.root.1 inputdata/recast_daod.root
Providing files to yadage
Since we have a signal DAOD file for yadage to process, we’ll need a way to tell the yadage-run command where to look for the DOAD file. This functionality is provided by the
-d initdir=option. For example, if the file to input is namedinputfile.txt, and it’s located in the directoryinputdatasyntax for passing it to yadage-run as the variable inputfile would be:yadage-run workdir workflow.yml -p inputfile=inputfile.txt -d initdir=$PWD/inputdata
Exercise (10 min)
Part 1:
Fill in the FIXMEs in the following skeleton code to encode the first skimming step of the analysis in your steps.yml file.
skimming_step: process: process_type: interpolated-script-cmd script: | # Source the ATLAS environment [FIXME] # Run the AnalysisPayload executable to produce the output ROOT file, looping over **all** events. [FIXME: source setup script to run executables] [FIXME] {input_file} {output_file} environment: environment_type: docker-encapsulated image: [FIXME (use your gitlab registry image!)] imagetag: master publisher: publisher_type: interpolated-pub publish: selected_events: [FIXME]Part 2:
Fill in the following skeleton code to encode the corresponding workflow stage in your workflow.yml file.
stages: - name: skimming_step dependencies: [FIXME] scheduler: scheduler_type: singlestep-stage parameters: input_file: {[FIXME], output: signal_daod} [FIXME]: '{workdir}/selected.root' step: [FIXME]Solution
Part 1
skimming_step: process: process_type: interpolated-script-cmd script: | # Source the ATLAS environment source /home/atlas/release_setup.sh # Run the AnalysisPayload executable to produce the output ROOT file, looping over **all** events. source /Tutorial/build/x86_64-centos7-gcc8-opt/setup.sh AnalysisPayload {input_file} {output_file} environment: environment_type: docker-encapsulated image: gitlab-registry.cern.ch/[your_username (FILL IN!!!!)]/recast-standalone imagetag: master publisher: publisher_type: interpolated-pub publish: selected_events: '{output_file}'Part 2
stages: - name: skimming_step dependencies: [init] scheduler: scheduler_type: singlestep-stage parameters: input_file: {step: init, output: signal_daod} output_file: '{workdir}/selected.root' step: {$ref: steps.yml#/skimming_step}
Debugging Hints
Could temporarily reduce the number of events you run over with
AnalysisPayloadfor the purposes of debugging. This is done by supplying a third argument to theAnalysisPayloadexecutable specifying the number of events to run over.Remember you can use the
packtivity-validatecommand in your yadage container to quickly check if your step definition is valid:packtivity-validate steps.yml#/skimming_step
- The step can be tested in your yadage container with a
packtivity-runcommand like:packtivity-run steps.yml#/skimming_step -p input_file="'{workdir}/inputdata/recast_daod.root'" -p output_file="'{workdir}/selected.root'"
- The workflow can be tested with a
yadage-runcommand like:yadage-run workdir workflow.yml -p signal_daod=recast_daod.root -d initdir=$PWD/inputdata- The output file should be located under
workdir/skimming_step/selected.rootafter a successfulyadage-run.
Key Points
The yadage syntax for defining our analysis steps is not too different from our helloworld example.
The main things that change are the details of the script processes and the docker image to run.